Use Case 1 HOW TO FIND NEGATIVE REGULATORS OF A SPECIFIC GENE
Biological question
Which Transcription Factors (TFs) and microRNAs (miRs) inhibit PTEN and can be investigated as a cancer therapy target?
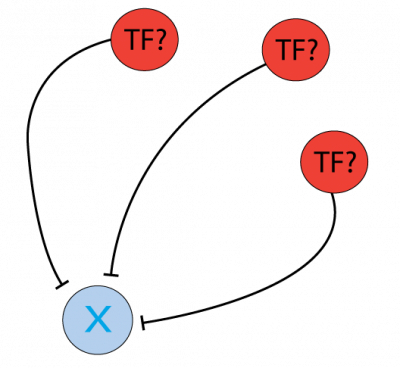
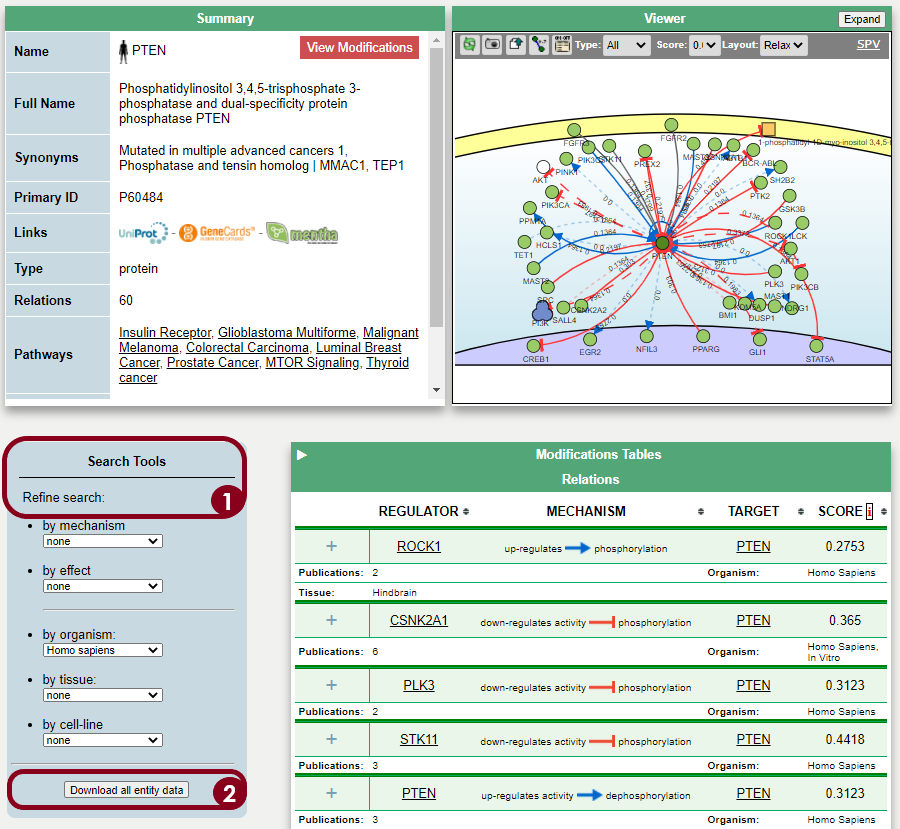
The protein PTEN is encoded by a tumor suppressor gene that is mutated in a large number of cancers, for example, head and neck (PMID: 11801303) and glioma cancer (PMID: 12085208). Regulation of the protein level can be done at the gene transcription level, by Transcription Factors, or post-transcriptionally, for instance by miRNA inhibition. To find the regulators at both levels, the Cytoscape software can be used as a platform to display the information of different resources and databases on this matter. General instructions about how to use this software and the software apps are available at //cytoscape.org/
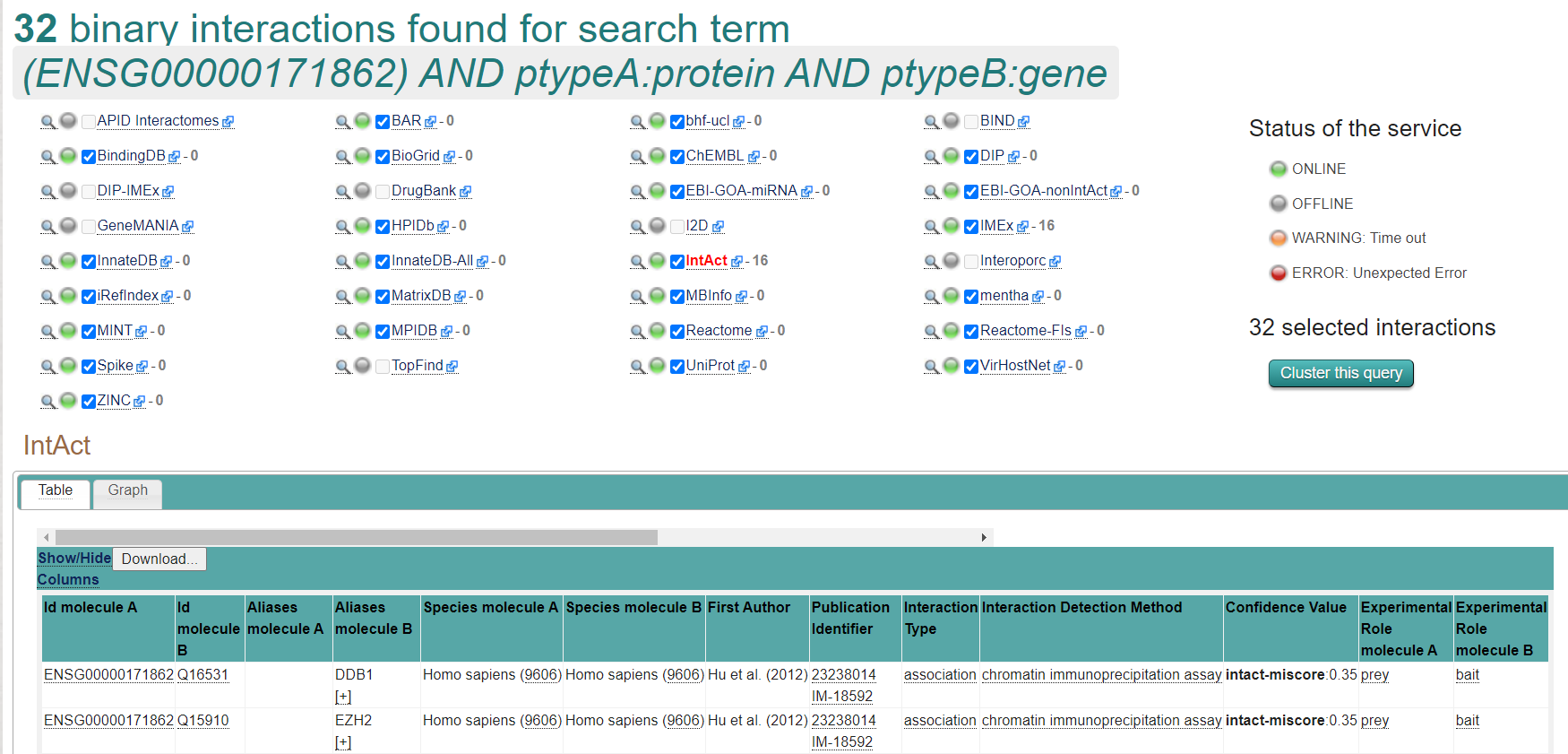
- To find all interactions with PTEN and genes, paste the following query into IntAct or PSICQUIC (adapt to your protein of interest): (ENSG00000171862) AND ptypeA:protein AND ptypeB:gene
- To find all interactions with PTEN mRNA and miRNAs, paste into IntAct or PSICQUIC: (ENST00000371953) AND ptypeA:mRNA AND ptypeB:mirna

- PTEN IDs needed to be included in the analysis: ENSG00000171862 ENST00000371953 P60484 PTEN
- The network titles can be edited and the different networks can be merged with the Cytoscape functions. The default style can be changed to the PSI-MI 2.5 Style for better interpretation.
Results from PSICQUIC View

Direct PSICQUIC query in Cytoscape
ADDITIONAL ANALYSIS
One thing that can be done to add the causality relationship information into the Cytoscape network is to use the SIGNOR database. The interactions already present in the query network might be described in SIGNOR, and, if so, these annotations can be added in Cytoscape to provide directionality in the network.